Galaxy is massive (if you’ll forgive the pun), with hundreds of tutorials, thousands of available tools, tens of thousands of active users, and hundreds of thousands of jobs executed by those users, on servers spread around the world. There are also a large number of options to use Galaxy. In this post, we will take a look at the currently available options, benefits, and tradeoffs for each. If you would like to watch a webinar on this topic, check out this recording.
Which Galaxy to use based on user role
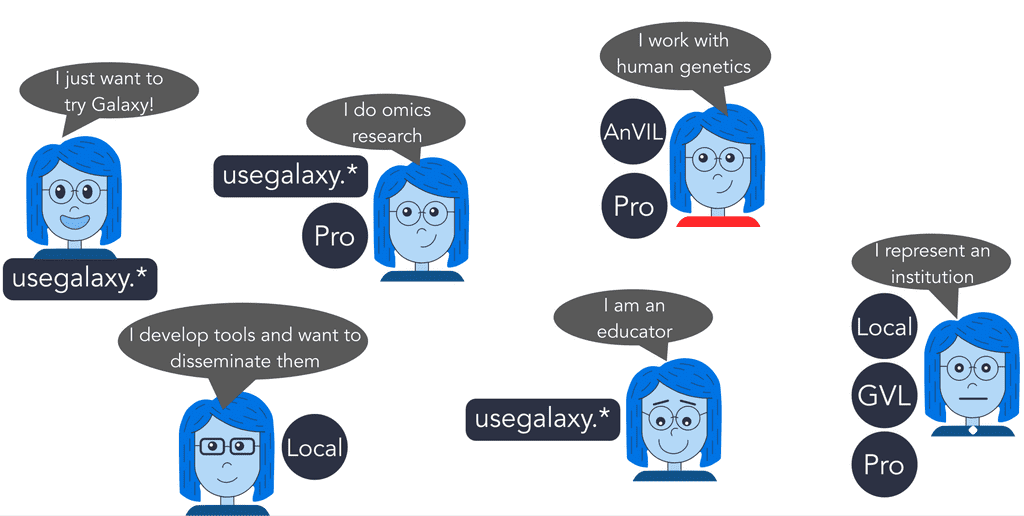
If you are a student or researcher without IT support and are running into limitations with local infrastructure with the Cloud being too technical, use one of the free usegalaxy.* servers.
If you are a researcher working with protected datasets, or if you are working with public data that is too large to analyze locally, or have access-controlled data with special requirements, use Galaxy on AnVIL or Galaxy Pro.
If you are a researcher working with large datasets (>250GB), have concerns about data privacy, or need high-performance hardware, use a local installation or Galaxy Pro.
Which Galaxy to use based on use case
If you want to quickly try Galaxy or complete a training session, use one of the free usegalaxy.* servers.
If you do omics research, depending on the size of your data and processing requirements, use either one of the usegalaxy.* servers or Galaxy Pro.
If you work with human genetics, use AnVIL or Galaxy Pro.
If you develop tools and want to disseminate them, use a local installation of Galaxy.
If you are an educator, use one of the usegalaxy.* servers.
If you represent a group or an institution and want to provide Galaxy for multiple users, use either a local installation of Galaxy, Galaxy Pro, or the Genomics Virtual Lab.
Digestible dive into each option for using Galaxy
Let’s now take a look at each of the mentioned options in more detail to see what exactly each option offers and how you go about accessing those. Feel free to also check out the Galaxy Platform Directory that will always have the most up-to-date list.
usegalaxy.*
Usegalaxy.* is a set of public servers hosted for free on national infrastructures around the world. There are half a dozen such servers available with more planned, including usegalaxy.org (aka Main), usegalaxy.eu, usegalaxy.org.au, usegalaxy.fr, usegalaxy.no, usegalaxy.be. Collectively, these servers offer thousands of tools, latest reference data, ability to share and collaborate with others, and powerful computing infrastructure. Each server is independent so you would need to register on each separately and any data you upload or analyze is local to the particular server. Furthermore, each server comes with different quotas in terms of how much storage and computing is available and a fixed toolset. It is also important to note that none of these servers are suitable for uploading protected data. Most of the training tutorials have been validated to run end-to-end on these servers so they are a phenomenal learning platform. Being shared servers, using these for workshops and training may not be ideal because of the variable queue wait times due to server contention. One option is to reach out to the project outreach and leverage the TIaaS framework to accommodate such scenarios.
An extension of the usegalaxy.* servers are the Galaxy Public servers. This is a list of 100+ servers that are accessible to any academic researcher in the world for free. Each of the individual servers offer a specific toolset for a specific type of analysis or domain, such as climate science tools or natural language processing among many different omics-inspired ones. These servers may have usage quotas. For the current list, check out the Public Servers section of the Galaxy Platform Directory.
AnVIL
AnVIL is a cloud platform that provides a unified computing environment for hosting analysis applications and large NIH NHGRI datasets, including genomics datasets, phenotypes, and metadata (eg, CCDG, CMG, GTEx,1000G, eMerge). This environment offers security assurances to allow operations on protected data (eg, dbGaP data), and the cloud environment is scalable to accommodate the needs of different types of analyses. Galaxy is one of the flagship applications available in AnVIL allowing researchers to readily launch their own instance of Galaxy and install any missing tools. For the time being, there are some usability limitations that will be resolved with time, including limited storage space and processing capacity. Using Galaxy on AnVIL will incur costs proportional to the usage. To get started with AnVIL, visit anvilproject.org.
Local installation
Galaxy is an open source software application licensed under the Academic Free License (AFL) v3.0. In turn, Galaxy can be installed on local infrastructure ranging from a laptop to a HPC cluster. Local installations of Galaxy are suitable for scenarios where you want to control user quotas, use custom tools, work on private data, or develop Galaxy tools. This mode of using Galaxy offers the most flexibility and control but it does require local infrastructure proportional to the workload. A Galaxy installation actively used by 10 or more users will require a near full-time system administrator to manage it. There are multiple ways to install Galaxy and which method is used is dependent on the available expertise, infrastructure, and intended usage. We recommend to use the Galaxy Helm chart and deploy Galaxy on Kubernetes as the most feature-full and robust deployment mechanism.
Galaxy Pro
Galaxy Pro is a subscription-based, managed installation of Galaxy where we handle all aspects of installing and managing the software and hardware while you can readily use Galaxy without quotas or system administration. Galaxy Pro Custom is a product line that offers maximum flexibility with options such as custom tools, custom tool panel, integration with your data sources (eg, S3 buckets, sequencers), complete infrastructure isolation for enhanced privacy, no quotas or queue wait time, support from an application scientist, and more. We are also piloting a general purpose Galaxy Pro Researcher product, which offers a common (but evolving) toolset, no queue wait times, and no quotas at a significantly lower price point than the Custom offering. The Researcher product will be generally available later this year. For more information, check out our Product page or contact us to request a demo.
Genomics Virtual Lab (GVL)
The GVL is a ready-made installation of Galaxy for the cloud. The GVL can be launched on a number of cloud providers (AWS, GCP, OpenStack) with software handling the infrastructure management and installation of Galaxy. With the GVL, the cloud infrastructure can readily scale to accommodate variable processing and storage requirements. The GVL can also be used to install a custom toolset or operate on private data. The cost of using the GVL is proportional to the workload being processed and all billing is handled through the chosen cloud provider. For academic researchers, the GVL can be free via one of the national cloud providers, such as Jetstream and NeCTAR. Because of the complexity of the software stack used by the GVL, it is highly recommended it be set up and actively managed by a system administrator for his/her users.
Conclusions
There you have it, the various ways one can use Galaxy in a digestible format. The bottom line is that regardless whether you are starting out in genomics and looking for a platform you can access right now, working with large human datasets and are looking for a system that can scale, or working with analyses ranging from RNA-Seq to drug discovery, there is an option out there to do it with Galaxy. It is worth noting that the available Galaxy services are constantly evolving so we will periodically be updating this post to keep it current. Check back in.